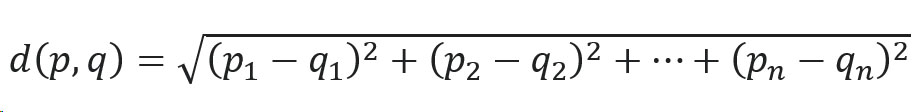All data can be vectorized – words, phrases, images, audio and documents. Vector databases are a tool which for most of the cases implements some kind of algorithm for approximating nearest neighbor which happens to be useful for similarity search between images or sounds and text, multiple contexts searching engines and LLMs improving domain-specific responses of large language models. When user provides the prompt, the feature vector is extracted, and the database is queried to retrieve the most relevant documents which then are added to the context of LLM and it creates a response to the prompt in this context. One of the most popular recently of the approximating nearest neighbor algorithms are Hierarchical Navigable Small World (HNSW) graphs which is one of the best performers in the benchmarks.
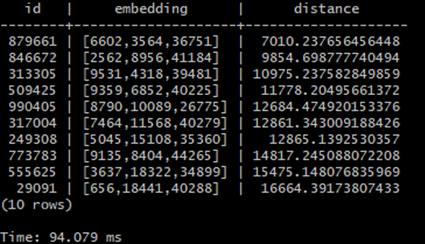
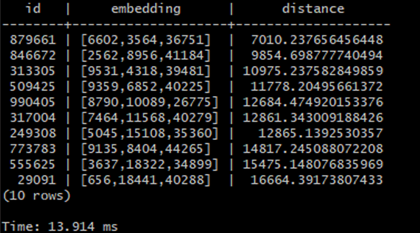
In this article I’ll try to familiarize you with this concept based on a simple example created using PostgreSQL plugin pgvector which introduces tools for storing, indexing and querying vectors inside postgres database and I will also show you how the indexing impacts the time of queries for the vectors.